Posted: June 2017
By Νiki Karachaliou, MD, PhD
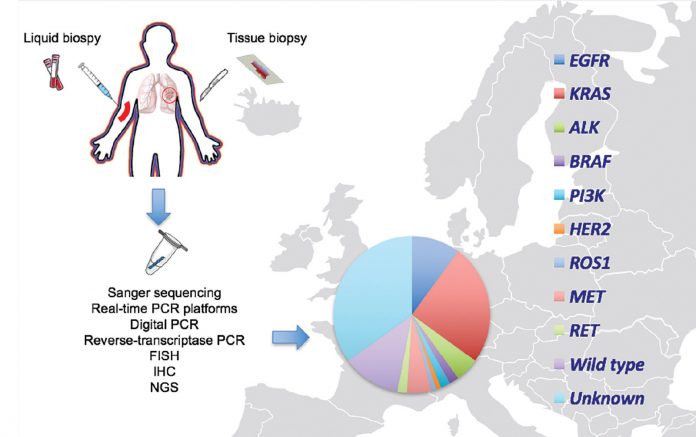
The identification of oncogenic driver alterations in non-small cell lung cancer (NSCLC), such as EGFR mutations and ALK rearrangements, each of which confers sensitivity to small tyrosine kinase inhibitors, has made EGFR and ALK testing a necessity in routine molecular pathologic diagnosis. Furthermore, next-generation sequencing (NGS) studies have divided NSCLC into molecular subtypes defined by distinct somatic alterations,1,2 which have led to an increasing interest in identifying additional targetable alterations in this disease.3 EGFR mutation testing is routinely available in 70% of the population worldwide, but it remains costly at a rate of $500 or more in the majority.4 Various techniques like conventional Sanger sequencing, real-time PCR platforms, digital PCR, and NGS are used to detect EGFR mutations. Fluorescence in situ hybridization (FISH), immunohistochemistry (IHC) reverse-transcriptase PCR, and some forms of NGS platforms are used for ALK gene fusion detection (Figure). Following the results of the KEYNOTE-024 trial that led to the FDA approval of pembrolizumab for first-line therapy in patients with PD-L1 expression of 50% or higher PD-L1 testing by IHC has become another very important biomarker.5 But it remains unclear which is best: PD-L1 testing, or PD-L1 plus other markers (e.g., tumor mutation burden, or neoepitopes,6 or IFN-γ signatures7), to enhance response prediction and selection of patients for anti-PD1/ L1 therapies.
In 2009, the Spanish Lung Cancer Group demonstrated that a large-scale screening study for EGFR mutations was feasible and reported a prevalence of 16.6% among Spanish patients with NSCLC.8 As a result, the Spanish Association of Medical Oncology and the Spanish Association of Pathology recommend routine EGFR mutation and ALK rearrangement testing in all NSCLC of nonsquamous cell subtype, or nonsmokers regardless of histologic subtype. The analyses should be performed in laboratories participating in external quality control programs, and the results should be provided no more than a week after the pathologic diagnosis.9 To this end, a nationwide platform funded by AstraZeneca was implemented in Spain for large-scale screening of EGFR mutations in the tissue and blood of patients with advanced NSCLC. Routine testing for other molecular abnormalities is not considered necessary in current clinical practice. In our molecular diagnostic laboratory, EGFR deletions in exon 19 and exon 21 point mutations in codon 858 are examined in tissue and blood with a 5′ nuclease PCR assay in the presence of a protein nucleic acid clamp, designed to inhibit the amplification of the wild-type allele.10 The analysis of the gatekeeper T790M mutation in exon 20 is always included in both pre-treatment11,12 and post-treatment13 tumor samples.
The Network Genomic Medicine (NGM) in Cologne, Germany, was the first group to screen for genomic alterations in all histological subtypes of lung cancer.14 Since then, the NGM has made great progress, implementing genotyping by NGS and genomic-driven treatment trials (e.g., EUCROSS: NCT02183870).15 From April 2012 to April 2013, the French Cooperative Thoracic Intergroup (IFCT) screened 17,664 patients with advanced NSCLC for oncogenic drivers through a nationwide program funded by the French National Cancer Institute.16 EGFR mutations and ALK rearrangements, as well as ERBB2, KRAS, BRAF, and PIK3CA mutations were assessed either concurrently or with a sequential approach in 28 certified regional genetics centers in France, using the Sanger sequencing method or a more sensitive validated allele-specific technique.16,17 The Lung Cancer Mutation Consortium (LCMC) initiative clearly demonstrated a clinical benefit for patients who are molecularly profiled and receive a matched targeted agent.16
According to the consensus of the Italian Association of Medical Oncology and the Italian Association of Pathology and Cytopathology, all patients with NSCLC of nonsquamous cell subtype should be tested for EGFR and KRAS mutations within 2 to 5 days after the pathologic diagnosis. Negative cases should be prescreened for elevated ALK protein by IHC before final confirmation by FISH.18 Through the Alliance Against Cancer,19 a national molecular screening program evaluating the use of NGS in patients with advanced NSCLC is going to be initiated in Italy by June 2017.
The Swiss Lung Pathology Group recommends gene sequencing analysis (through various NGS platforms) for EGFR, KRAS, BRAF, and HER2 mutations, as well as prescreening for elevated ALK and ROS1 protein by IHC (positive cases should be confirmed by FISH). For negative cases, sequential testing for MET amplification or RET rearrangements is highly recommended, as well as the evaluation of PD-L1 protein expression, MET exon 14 skipping mutations, and NTRK1 rearrangements.
The above mentioned molecular diagnostic algorithms represent a shift in lung cancer diagnosis and treatment, but at the same time pose several challenges for Europe and beyond. For instance, there is a paradox between the development of minimally invasive techniques, resulting in small tissue samples and the need to obtain large enough samples for the analyses of a growing number of biomarkers. High-quality diagnostic samples, molecular profiles of various samples (including plasma genotyping), new sampling procedures, and high-sensitivity tests should all be combined to provide great amounts of information from increasingly smaller amounts of tissue. A Spanish Lung Liquid versus Invasive Biopsy Program (SLLIP) is currently running in Spain, with the primary objective of demonstrating the non-inferiority of cell-free circulating tumor DNA (cfDNA)-based versus tumor tissue-based genotyping as it pertains to the detection of clinically actionable biomarkers in first-line, treatmentnaive, metastatic nonsquamous NSCLC. The Guardant360 cfDNA-targeted NGS panel is used for the cfDNA-based genotyping in the SLIPP study. The logistical challenges of implementing molecular diagnostics in clinical practice, including the access to targeted therapies, are also important issues.
In the United Kingdom, NGS is applied to samples from patients with advanced NSCLC to screen for clinically actionable known drivers, through the Cancer Research UK Stratified Medicine Program 2 (SMP2).20 According to the results obtained, patients are recruited to The National Lung Matrix Trial (NCT02664935), a phase II umbrella study.21 A similar umbrella study is ongoing in France: SAFIR02 Lung trial (NCT02117167).22 Even with these initiatives for matching drugs to tumor profiles, the goals are not always reached: many patients are not able to go through a successful biopsy, actionable targets are detected in less than half of the patients, and only a minority of patients are finally treated with targeted therapies. A joint European strategy for NGS sequencingbased molecular diagnostics of lung cancer will definitively establish a database for the evaluation of personalized lung cancer therapy, increase access to new drugs, and develop models for reimbursement adapted to diverse national health care systems.
References
1. Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511(7511):543-550.
2. Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012; 489(7417):519-525.
3. Hunter DJ. Uncertainty in the Era of Precision Medicine. N Engl J Med. 2016; 375(8):711-713.
4. Carbonnaux M, Souquet PJ, Meert AP, Scherpereel A, Peters M and Couraud S. Inequalities in lung cancer: a world of EGFR. Eur Respir J. 2016; 47(5):1502-1509.
5. Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1- Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016; 375(19):1823-1833.
6. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015; 348(6230):124-128.
7. Ayers M, Lunceford J, Nebozhyn M. Relationship between immune gene signatures and clinical response to PD-1 blockade with pembrolizumab (MK-3475) in patients with advanced solid tumors. J Immunother Cancer. 2015; 3(Suppl 2):P80-P80.
8. Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009; 361(10):958-967.
9. López-Ríos F, de Castro J, Concha Á, et al. Actualización de las recomendaciones para la determinación de biomarcadores en el carcinoma de pulmón avanzado de célula no pequeña. Consenso Nacional de la Sociedad Española de Anatomía Patológica y de la Sociedad Española de Oncología Médica. Rev Esp Patol. 2015; 48(2):80-89.
10. Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol. 2015; 1(2):149-157.
11. Rosell R, Dafni U, Felip E, C, et al. Erlotinib and bevacizumab in patients with advanced non-smallcell lung cancer and activating EGFR mutations (BELIEF): an international, multicentre, single-arm, phase 2 trial. Lancet Respir Med. 2017.
12. Karachaliou N, Morales-Espinosa D, Molina Vila MA, et al. P2.06-010 AZD9291 as 1st-Line Therapy for EGFR Mutant NSCLC Patients with Concomitant Pretreatment EGFR T790M Mutation. The AZENT Study. J Thorac Oncol. 12(1):S1074-S1075.
13. Mok TS, Wu YL, Ahn MJ, , et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med. 2017; 376(7):629-640.
14. Clinical Lung Cancer Genome P and Network Genomic M. A genomics-based classification of human lung tumors. Sci Transl Med. 2013; 5(209):209ra153.
15. Michels S, Gardizi M, Schmalz P, T, et al. MA07.05 EUCROSS: A European Phase II Trial of Crizotinib in Advanced Adenocarcinoma of the Lung Harboring ROS1 Rearrangements-Preliminary Results. J Thorac Oncol. 12(1):S379-S380.
16. Barlesi F, Mazieres J, Merlio JP, , et al. Routine molecular profiling of patients with advanced nonsmall- cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet. 2016; 387(10026): 1415-1426.
17. Rosell R and Karachaliou N. Large-scale screening for somatic mutations in lung cancer. Lancet. 2016; 387(10026):1354-1356.
18. Marchetti A, Ardizzoni A, Papotti M, et al. Recommendations for the analysis of ALK gene rearrangements in non-small-cell lung cancer: a consensus of the Italian Association of Medical Oncology and the Italian Society of Pathology and Cytopathology. J Thorac Oncol. 2013; 8(3):352-358.
19. De Paoli P, Ciliberto G, Ferrarini M, et al. Alliance Against Cancer, the network of Italian cancer centers bridging research and care. J Transl Med. 2015; 13:360.
20. Hiley CT, Le Quesne J, Santis G, et al. Challenges in molecular testing in non-small-cell lung cancer patients with advanced disease. Lancet. 2016; 388(10048):1002-1011.
21. Middleton G, Crack LR, Popat S, et al. The National Lung Matrix Trial: translating the biology of stratification in advanced non-small-cell lung cancer. Ann Oncol. 2015; 26(12):2464-2469.
22. Biankin AV, Piantadosi S Hollingsworth SJ. Patientcentric trials for therapeutic development in precision oncology. Nature. 2015; 526(7573): 361-370.