Permission granted by Dr. Marina Garassino
By Lynette M. Sholl, MD, and John W. Longshore, PhD, FACMG
Posted: February 12, 2020


The profound impact of immunotherapy in patients with lung cancer has been described extensively by many experts within the IASLC.1,2 There are, however, broad disparities in outcomes for patients with lung cancer treated with various anti‒PD-1/‒PD-L1 therapies, reflecting the tremendous heterogeneity of the disease itself. The PD-1/PD-L1 axis is the target of many approved immunotherapeutics, and consequently, the predictive power of PD-L1 expression on tumor cells has served as a major focus of clinical trials. Across these clinical trial and cohort studies, PD-L1 expression on tumor cells enriches for response to these therapies; however, its predictive ability is modest at best and is relevant only for certain inhibitors in specific contexts. This quagmire has led to the search for alternative or complementary biomarkers of response to immunotherapies.
Many lung cancers harbor large numbers of mutations resulting from extrinsic DNA damage from exposures like tobacco smoke or intrinsic DNA damage processes such as apolipoprotein B mRNA editing enzyme, catalytic polypeptide- like (APOBEC)-induced cytidine deamination.3,4 Correlative studies have consistently shown a relationship between high mutational load (or tumor mutation burden [TMB]) and response to immunotherapies. Mechanistically, this relationship is thought to be driven by the greater likelihood of generating immunogenic novel proteins (neoantigens) in highly mutated tumor cells. Whole-exome sequencing delivers an absolute quantitation of TMB (in number of mutations per exome); however, large-panel sequencing is more readily available in clinical practice. In general, the TMB can be extrapolated via bioinformatic methods from the focused data generated by the panel and reported as the number of mutations per megabase of genome sequenced.
Finding Answers for a Long List of Questions
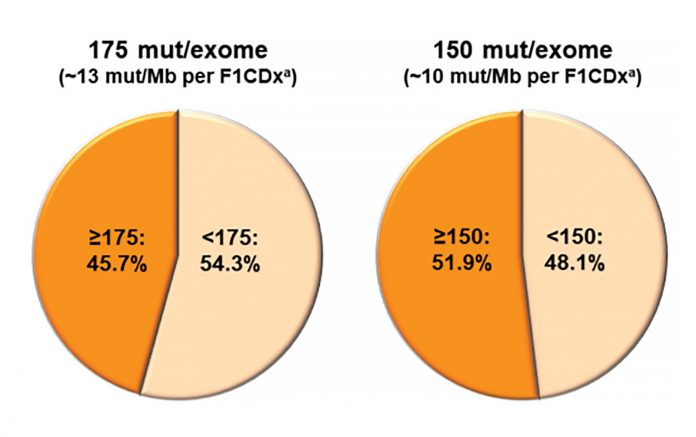
Although there is little doubt that TMB constitutes a unique characteristic of individual lung cancers, many questions remain about its tractability as a biomarker of response to immunotherapy. Given the wide range of possible TMB scores, a robust cutpoint optimized for sensitive and specific prediction of therapeutic response must be derived from clinical outcomes data. Proposed cutpoints have ranged from 10 to 15 mutations/Mb in tissue and six to 20 mutations/Mb in plasma-derived cell free (cf) DNA.5-9 Adding further complexity, “eligible” mutations have not been fully established—these may or may not include missense (nonsynonymous), synonymous, insertion-deletion mutations, and splice site mutations. Subclonal mutations—those present only in a subpopulation of tumor cells— may be included in some calculations but not others. Some panels require parallel sequencing of a paired normal specimen to exclude germline variants from analysis; others remove germline variants from tumor-only sequencing results using population database filters. The targeted content of panel next-generation sequencing itself may be a confounding factor, as certain genes are more or less prone to mutation and, thus, may bias the TMB estimation of a given panel. To address the disparities inherent to different panel designs and bioinformatic rules for variant calling, multiple investigators have undertaken systematic evaluation of TMB across different sequencing panels. Garrido-Martin and colleagues10 describe similar rates of “TMBhigh” calling at different cutpoints between three commercially available sequencing panels. However, Budczies and colleagues11 uncovered a substantial rate of TMB misclassification around 10 mutations/Mb using panels covering up to 1.4Mb of genomic content. They emphasized the need for a larger panel to more accurately assess TMB levels and proposed a three-tiered reporting approach to reduce misclassification. Applying whole-exome sequencing as a gold standard, Vokes and colleagues12 compared tumor-only and paired tumornormal sequencing approaches and proposed a transformation step to harmonize TMB results across specific panels. Larger multi-institutional efforts led by the Friends of Cancer Research and the Qualitätssicherungs-Initiative Pathologie have undertaken a multipronged approach examining bioinformatic and wet-lab variables influencing TMB calculation, with the ultimate goal of generating a global standard to permit cross-laboratory harmonization.13
Efforts to develop TMB as a robust biomarker are motivated by data suggesting that TMB can predict improvements in response and progression- free survival following immunotherapy. The data from trials are contradictory, however, with little evidence to date that higher TMB predicts better overall survival.14 In patients receiving combined PD-1 inhibition and chemotherapy, TMB does not appear to predict outcomes.15 In contrast, TMB may be a robust biomarker of response with PD-1 inhibitor monotherapy in the first and second lines in patients with PD-L1‒positive tumors.16 Ultimately, several questions must be addressed before TMB can be fully incorporated into clinical practice. In what clinical context will TMB best inform choice of therapy? What TMB cutpoint provides optimal sensitivity and specificity for response? Is tissue-based TMB required, or is cfDNA sufficient for use as a surrogate for TMB analysis? What type and size of targeted panel is required to generate a reliable TMB? How should TMB be considered in the context of other established or putative biomarkers including PD-L1 expression, immune environment, and genomic context?17,18 All of these issues are under intense scrutiny in the lung cancer research community. The IASLC Pathology Committee is working to summarize the existing issues and provide clarity on the potential role for TMB in the management of patients with lung cancer. ✦
About the Authors: Dr. Sholl is an associate professor of Pathology at Harvard Medical School and an Associate Pathologist in the department of Pathology at Brigham And Women’s Hospital. Dr. Longshore is director of Molecular Pathology at Carolinas Pathology Group, Atrium Health, Carolinas HealthCare System. Drs. Sholl and Longshore co-chair the IASLC Pathology Committee Molecular Pathology Working Group. Members of the working group are preparing a multidisciplinary review and perspective piece on the clinical utility of TMB and technical complexity of this biomarker.
References:
1. Garon EB, Hellmann MD, Rizvi NA, et al. Five-Year Overall Survival for Patients With Advanced NonSmall-Cell Lung Cancer Treated With Pembrolizumab: Results From the Phase I KEYNOTE-001 Study. J Clin Oncol. 2019;37(28):2518-2527.
2. Antonia SJ, Borghaei H, Ramalingam SS, et al. Four-year survival with nivolumab in patients with previously treated advanced non-small-cell lung cancer: a pooled analysis. Lancet Oncol. 2019;20(10):1395-1408.
3. McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463-1469.
4. Wang S, Jia M, He Z, Liu XS. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018;37(29):3924-3936.
5. Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med. 2018;378(22):2093- 2104.
6. Garassino MC. KEYNOTE 189: Tumor Mutational Burden Not Significantly Associated with Efficacy of Pembrolizumab (abstract). J Thorac Oncol. 2019.
7. Peters S. Abstract CT074: Tumor mutational burden (TMB) as a biomarker of survival in metastatic non-small cell lung cancer (mNSCLC): Blood and tissue TMB analysis from MYSTIC, a Phase III study of first-line durvalumab ± tremelimumab vs chemotherapy. Cancer Res. 2019;79(13).
8. Wang Z, Duan J, Cai S, et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019;5(5):696-702.
9. Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441-1448.
10. Garrido-Martin EM, Ramos- Paradas J, Hernandez Prieto S, et al. LBA17Harmonization study of tumour mutational burden determination in non-small cell lung cancer (NSCLC). Ann Oncol. 2019;30(Suppl_5).
11. Budczies J, Allgauer M, Litchfield K, et al. Optimizing panel-based tumor mutational burden (TMB) measurement. Ann Oncol. 2019;30(9):1496-1506.
12. Vokes NI, Liu D, Ricciuti B, et al. Harmonization of Tumor Mutational Burden Quantifi cation and Association With Response to Immune Checkpoint Blockade in Non–Small-Cell Lung Cancer. JCO Precis Oncol. 2019;1-12.
13. Stenzinger A, Allen JD, Maas J, et al. Tumor mutational burden standardization initiatives: Recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes Chromosomes Cancer. 2019;58(8):578- 588.
14. Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30(5):863-870.
15. Paz-Ares L, Langer CJ, Novello S, et al. LBA80Pembrolizumab (pembro) plus platinum-based chemotherapy (chemo) for metastatic NSCLC: Tissue TMB (tTMB) and outcomes in KEYNOTE-021, 189, and 407. Ann Oncol. 2019;30.
16. Herbst RS, Lopes G, Kowalski DM, et al. LBA79Association between tissue TMB (tTMB) and clinical outcomes with pembrolizumab monotherapy (pembro) in PD-L1-positive advanced NSCLC in the KEYNOTE-010 and -042 trials. Ann Oncol. 2019;30.
17. Lu S, Stein JE, Rimm DL, et al. Comparison of Biomarker Modalities for Predicting Response to PD-1/PD-L1 Checkpoint Blockade: A Systematic Review and Meta-analysis. JAMA Oncol. 2019 Jul 18. [Epub ahead of print].
18. Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018;8(7):822-835.