By Christian Rolfo, MD, PhD, MBA, Dr.h.c., and Lori Alexander, MTPW, ELS, MWC
Posted: August 14, 2019
Targeted therapy has become the standard of care in patients with advanced NSCLC and oncogenic drivers, and tissue biopsy has emerged as the gold standard in the molecular diagnosis of the disease. Guidelines recommend that all patients with newly diagnosed advanced NSCLC have molecular testing to detect five to eight predictive biomarker mutations,1-3 and as the number of targetable oncogenic alterations in NSCLC continues to grow, multiplexed genetic sequencing panels are preferable over sequential or multiple singlegene tests.2 However, in most patients, lung cancer is diagnosed on the basis of small tissue biopsy or cytologic samples; consequently, complete molecular testing is not achieved in a substantial proportion of cases. Studies have shown that the use of molecular testing varies widely across practices around the world4,5 and that complete genotyping is carried out in approximately 8% of patients with newly diagnosed advanced NSCLC in the United States.6
Over the past few years, liquid biopsy has generated great interest as a minimally invasive diagnostic assay to overcome the challenges of tissue biopsy and thus increase the number of patients who can be tested for druggable biomarkers and can receive appropriate treatment based on complete molecular information. In addition, the time to the start of treatment is delayed by a long turnaround time between receipt of a tissue sample and reporting of relevant results; this delay has often led to the initiation of chemotherapy before genomic results are available to avoid substantial deterioration in the patient’s clinical condition.4,6,7 Liquid biopsy holds promise as a viable alternative to tissue biopsy and has been evaluated for multiple potential clinical uses, including biomarker identification, patient selection for treatment, early cancer detection, and drug resistance monitoring.
Guidelines Related to Liquid Biopsy
EGFR testing on cell-free DNA (cfDNA) is currently recommended in the IASLC/College of American Pathologists (CAP)/Association for Molecular Pathology (AMP) guidelines for patients with limited and/or insufficient tumor tissue for molecular testing; it has also been recommended to identify EGFR T790M mutations in EGFR-mutated NSCLC progressing after treatment with first- or second-generation EGFR tyrosine kinase inhibitors. Testing of a tumor sample is recommended if the results of liquid biopsy are negative.2 An increasing number of next-generation sequencing (NGS) platforms have been recently developed to not only improve the fidelity of the molecular analysis but also to increase the number of tests performed on a single specimen, allowing simultaneous evaluation of single-base variants, indels, copy number variations, and chromosomal rearrangements.4 However, high cost and limited availability restrict the widespread use of these platforms.
In 2018, an expert review conducted by the American Society of Clinical Oncology (ASCO) and CAP led to the conclusion that the evidence related to the use of liquid biopsy was insufficient to recommend its routine use for making treatment decisions and monitoring treatment.8 However, later that year, the IASLC published a statement paper to note that “liquid biopsy approaches have significant potential to improve patient care, and immediate implementation in the clinic is justified in a number of therapeutic settings relevant to NSCLC.”9 Several liquid biopsy assays are available for use in clinical practice, but only one is approved by the U.S. Food and Drug Administration (FDA): the cobas® EGFR Mutation Test (Roche, Basel, Switzerland) in NSCLC.10
Head-to-Head Comparison of Liquid and Tissue Biopsy
One issue of concern with liquid biopsy testing is how its results compare with those of tissue biopsy. Among the most recent studies in this area is the multicenter prospective Noninvasive versus Invasive Lung Evaluation (NILE) trial, which was conducted to determine whether a validated and highly sensitive plasma NGS test (Guardant360; Guardant Health, Redwood City) used at the time of diagnosis of NSCLC could prove noninferior to standard-of-care tissue genotyping in identifying guideline-recommended genomic biomarkers; it also set out to evaluate potential advantages of cfDNA testing.11 Tissue genotyping included NGS, polymerase chain reaction (PCR) hotspot testing, fluorescent in situ hybridization (FISH) and/ or immunohistochemistry (IHC), or Sanger sequencing, and the biomarkers included EGFR mutations, ALK fusions, ROS1 fusions, BRAF V600E mutation, RET fusions, MET amplification and MET exon 14 skipping variants, HER2 mutations, and KRAS mutations.
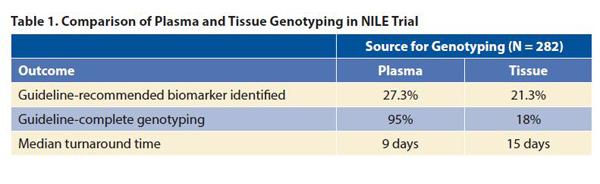
The results showed that testing of plasma with a 73-gene NGS at baseline was not inferior to standard-of-care tissue genotyping (p < 0.0001 for noninferiority; Table 1). For the four biomarkers with FDA-approved therapies, the concordance of liquid biopsy to tissue biopsy was greater than 98.2%, with 100% positive-predictive value. The use of cfDNA increased the number of patients with an identified guidelinerecommended biomarker by 48%, from 60 patients to 89, including those who had negative results on tissue genotyping (seven patients), those who did not have tissue genotyping (16 patients), and those for whom the amount of tissue was insufficient for testing (six patients). Liquid biopsy allowed guideline-complete genotyping in significantly more patients than tissue biopsy (p < 0.0001) and was associated with a significantly shorter median turnaround time (p < 0.0001; Table 1).
These results may lead to a change in the current diagnostic paradigm in advanced NSCLC, in which tissue genotyping is performed first and liquid biopsy is obtained only when tissue is not available for genomic testing, to one in which liquid biopsy moves upfront (a so-called blood-first approach) and tissue is reserved for IHC testing for PD-L1 and genotyping testing when the results of liquid biopsy testing are negative or inconclusive.
Novel Approach to Plasma Genotyping
One of the potential challenges in plasma genotyping is the identification of tumor-derived mutations of hematopoietic origin (due to a phenomenon called clonal hematopoiesis), generating false-positive results. This poses a major challenge when liquid biopsy is used to evaluate minimal residual disease and for early cancer detection,12 and it is a potential cause of discordance between tumor and plasma genotyping.13
Researchers for the Actionable Genome Consortium sought to investigate the role of an ultra-deep plasma NGS assay with clonal hematopoiesis filtering to guide the treatment of patients with NSCLC.14 The researchers used a novel approach that incorporated white blood cell sequencing to filter somatic mutations attributable to clonal hematopoiesis. With this approach ultra-deep NGS achieved overall high concordance with tissue testing across a variety of actionable oncogenes, with 75% sensitivity for de novo plasma detection of known oncogenic drivers in 68 of 91 cases and 100% specificity of plasma NGS for patients who had negative results for oncogenic drivers on tissue testing with NGS in 19 of 19 cases. Furthermore, plasma NGS allowed the identification of four oncogenic drivers among 17 patients in whom the status of oncogenic drivers was unknown because of insufficient tissue. The findings of orthogonal validation with plasma droplet digital PCR (ddPCR) for EGFR or KRAS mutations were nearly identical to those of plasma NGS in 21 of 22 patients, with only one driver mutation not detected by the NGS assay (this mutation had a low variant allele fraction of 0.04% by ddPCR).
Monitoring Response
The results of the IMMUNO-PREDICT trial were presented at the 2019 ASCO/Society of Immunotherapy for Cancer Clinical Immuno-Oncology Symposium.15 Using plasma NGS with tagged amplicon sequencing of hotspots and coding regions from 36 genes (Inivata; Granta Park, Cambridge, UK), Guibert et al. analyzed samples (collected at baseline and after 1 month of therapy) from 39 patients who had a response and 47 patients who did not have a response to second-line nivolumab. “Response” was defined as progression-free survival of longer than 6 months, and “no response” was defined as progressive disease at first evaluation. The presence of specific genetic alterations was evaluated according to outcomes. The presence of a targetable oncogenic driver (EGFR mutation or ALK fusion) was associated with primary resistance to immune checkpoint blockage, and the identification of a PTEN and/or STK11 mutation (b-PS(+)) correlated with a poorer outcome (median progression-free survival, 1.5 vs. 8 months, p = 0.0007) compared with b-PS(-). In contrast, KRAS and/or TP53 mutations (b-KP-Tv(+)) predicted improved outcome compared with b-KP-Tv(-) (median progression-free survival, 11 vs. 2 months, p = 0.0088).
Combining these results, the authors were able to develop an immune score: a low immune score was characterized by an oncogenic driver and/or b-PS(+) and/or b-KP-Tv(-), and a high immune score, by no driver, b-PS(-), and b-KPTv(+). Th e progression-free survival was significantly shorter for patients with a low immune score than for patients with a high immune score (Table 2). Furthermore, molecular response was correlated with a longer median progression- free survival (14 months for patients with an early decrease in circulating tumor DNA compared with 2 months for patients with increased levels of circulating tumor DNA; p < 0.0001; HR: 2.7); when a cut-off of 30% and 50% of plasma response was used, the ability of circulating tumor DNA to predict radiographic response increased (HR of 4.0 and 4.17, respectively).

These data further confirm the predictive role of STK11, KRAS, and TP53 mutations detectable in plasma on the activity of immune checkpoint inhibitors in NSCLC16,17 and support the potential use of NGS genotyping on circulating tumor DNA in the pretreatment evaluation of patients who are candidates for immunotherapy.
Patient Perspective
The advent of targeted therapy and immunotherapy in advanced NSCLC has led to a growing population of people living longer with the disease, which alters the implications of research and post-treatment surveillance. For example, issues such as long-term effects, which were once unimportant because of the short survival expectations, now must be evaluated. The patient perspective is essential to the discussion of genomic testing with liquid biopsy. Patient-reported outcomes after tissue and liquid biopsy testing should be compared. Cost is another important factor, as the high cost can be prohibitive; not all NGS testing is covered by Medicare or third-party insurers (lab-developed testing systems are typically not covered). Also, messaging about biomarkers must be consistent to avoid confusion about why biomarker testing is important, who should have testing, and when testing should be done.
The advent of targeted therapy and immunotherapy in advanced NSCLC has led to a growing population of people living longer with the disease, which alters the implications of research and post-treatment surveillance.
In its 2018 Post-Treatment Surveillance Workshop, the National Cancer Institute addressed the importance of patient perspectives, and Janet Freeman-Daily, a lung cancer advocate, spoke about the importance of the patient voice. She said that surveillance must be meaningful to patients, not just research, and she encouraged attendees to learn what matters most to their patients and to engage in true shared decision-making. ✦
About the Authors: Dr. Rolfo is the director of the Thoracic Medical Oncology and the Early Clinical Trials at the University of Maryland Marlene and Stewart Greenebaum Comprehensive Cancer Center. Ms. Alexander is a certified medical writer and the education director at the American Medical Writers Association.
References:
1. Ettinger DS, Wood DE, Aisner DL, et al. NCCN Clinical Practice Guidelines in Oncology. Non-small cell lung cancer, version 1.2019. www.nccn.org. Accessed May 22, 2019.
2. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018;13(3):323-358.
3. Kalemkerian GP, Narula N, Kennedy EB, Temin S, et al. Molecular testing for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors guideline endorsement. Systemic therapy for stage IV non-small-cell lung cancer: American Society of Clinical Oncology clinical practice guideline update. J Clin Oncol. 2018;36(9):911-918.
4. Ruggiero JE, Rughani J, Neiman J, et al. Real-world concordance of clinical practice with ASCO and NCCN guidelines for EGFR/ALK testing in aNSCLC. J Clin Oncol. 2017;35(no. 8_suppl):212.
5. Lee DH, Tsao MS, Kambartel KO, et al. Molecular testing and treatment patterns for patients with advanced non-small cell lung cancer: PIvOTAL observational study. PLoS One. 2018; 13(8):e0202865
6. Gutierrez ME, Choi K, Lanman RB, et al. Genomic profiling of advanced non-small cell lung cancer in community settings: gaps and opportunities. Clin Lung Cancer. 2017;18(6):651- 659.
7. Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol. 2015;26(7):1415-21.
8. Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36(6):1631-1641.
9. Rolfo C, Mack PC, Scaglotti GV, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018;13(9):1248-1268.
10. US Food & Drug Administration. FDA approves first blood test to detect gene mutation associated with non-small cell lung cancer. June 1, 2016. www.fda.gov/news-events/press-announcements/fda-approves-first-blood-test-detect-gene-mutation-associated-non-small-cell-lung-cancer. Accessed May 22, 2019.
11. Leighl NB, Page RD, Raymond VM, et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin Cancer Res. 2019 April 15 [Epub ahead of print].
12. Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC – challenges to implementing ctDNAbased screening and MRD detection. Nat Rev Clin Oncol. 2018;15(9):577-586.
13. Hu Y, Ulrich BC, Supplee J, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24(18):4437-4443.
14. Li BT, Janku F, Jung B, et al. Ultra-deep nextgeneration sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol. 2019;30(4):597-603.
15. Guibert N, Jones G, Beeler JF, et al. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced nonsmall cell lung cancer. meetinglibrary.asco.org/record/170358/abstract. Accessed May 25, 2019.
16. Biton J, Mansuet-Lupo A, Pécuchet N, et al. TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin Cancer Res. 2018;24(22):5710-5723.
17. Arbour KC, Jordan E, Kim HR, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2018;24(2):334-340.