By Rachel Rosenthal, PhD, MSc, and
Charles Swanton, FRCP, BSc, PhD
Posted: June 2018
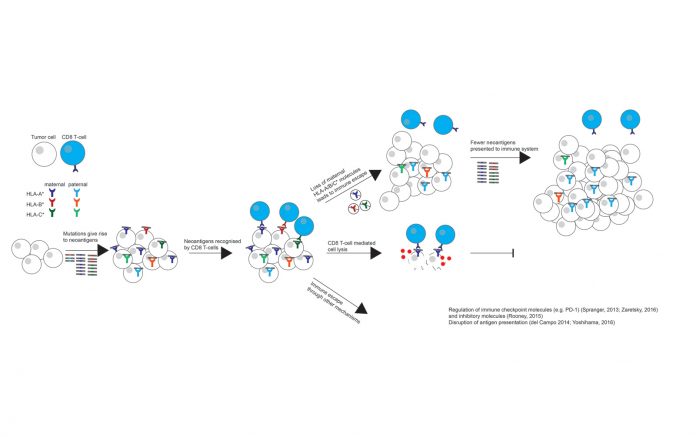
An evolving tumor and the immune system continuously adapt to each other. As a tumor develops increasing numbers of somatic alterations and disregulated genes, it must also find ways to avoid immune detection and elimination by activated immune cells.1 One route to withstand immune predation is through the disruption or prevention of antigen presentation, as reductions in antigen presentation can limit immune recognition. Indeed, a high proportion of cancer types have been found to acquire detrimental human leukocyte antigen (HLA) mutations,2 down-regulate HLA expression, 3-5 or abolish the function of the stabilizing molecule beta-2 microglobulin (B2M).6-8
Another means of HLA disruption is via loss of heterozygosity (LOH) at the HLA locus, wherein the maternal or paternal HLA haplotype is lost, impairing the immune system’s ability to recognize tumor antigens.9 This particular mechanism of immune evasion was recently documented in the case study of a patient who developed a resistant lesion after being treated with tumor-infiltrating lymphocytes composed of T-cell clones targeting KRAS G12D.10 The resistant lesion was found to have lost the HLA allele responsible for presenting the targeted neoantigen.
However, the polymorphic nature of the HLA locus has hampered the determination of copy number events affecting the locus, such as losses and amplifications, rendering a large-scale study of HLA LOH in human tumor samples and its effects on the tumor–immune system relationship infeasible. In a recently published study, we present a novel computational tool, Loss Of Heterozygosity in Human Leukocyte Antigen (LOHHLA), which can be used to determine HLA allele-specific copy number from sequencing data.11
To determine the prevalence of HLA LOH events in NSCLC (Fig.), we applied LOHHLA to the first published cohort from the Tracking Non–Small Cell Lung Cancer Evolution through Therapy (TRACERx) study. TRACERx is a multicenter, prospective cohort study, through which surgically resected NSCLC tumors are subject to high-depth, multi-region, whole-exome sequencing in order to investigate tumor evolution, intratumor heterogeneity, and the effects on clinical outcome.12
HLA LOH was identified in 40% of NSCLC samples. This was in contrast to other forms of HLA disruption, such as HLA mutations, which were only observed at a frequency of 3% in the TRACERx cohort. This observation suggests that HLA LOH may be a far more prevalent form of immune evasion. The multi-region aspect of the TRACERx dataset also allowed for the timing of HLA LOH events, as those that were identified in only a subset of tumor regions were subclonal in nature and occurred later in tumor evolution. Likewise, events that could be identified in every tumor region were considered early events in tumor evolution. Indeed, mapping specific HLA LOH events to tumor phylogenetic trees revealed that LOH at the HLA locus often occurred late in tumor evolution, on the branches of the phylogenetic tree. Strikingly, HLA LOH events sometimes mapped to multiple branches of the phylogenetic tree, suggesting that the loss had occurred at multiple time points over the course of a single tumor’s evolutionary history. A formal statistical analysis revealed that focal loss at the HLA locus occurred more frequently than expected by chance, suggesting strong selective pressure for the event late in tumor evolution, potentially in response to a shift in the equilibrium between immune recognition and evasion.
HLA LOH events were found to associate with a high subclonal mutation and increased APOBEC-mediated mutagenesis. Furthermore, and consistent with LOH at the HLA locus facilitating the accumulation of subclonal neoantigens, there was a significant enrichment for subclonal neoantigens predicted to bind to the lost HLA alleles as compared to the kept alleles, suggesting that disrupting HLA expression could be an effective mechanism of evading immune detection.
TRACERx tumors exhibiting LOH at the HLA locus also had increased PD-L1 positivity. Because the PD-L1 ligand binds to the inhibitory receptor PD-1, the expression of PD-L1 may reflect a response to an active immune system. Validation using expression data from NSCLC samples in The Cancer Genome Atlas confirmed that tumors harboring HLA LOH events had increased immune cell infiltration, reflective of an active immune microenvironment. These data are consistent with the hypothesis that HLA LOH facilitates immune escape later in tumor evolution in response to increased immune pressure.
Targeting neoantigens predicted to bind to HLA alleles already lost in the tumor may not effectively elicit a T-cell response. Furthermore, as HLA allele–specific loss has already once been observed in an immunotherapy-resistant lesion, it will be intriguing to investigate how frequently HLA LOH results in acquired immunotherapy resistance. Indeed, a recent publication has since investigated the HLA locus of patients treated with checkpoint blockade therapy and found that a subset of patients harboring an HLA LOH event had poorer survival.13


(Left to Right: Dr. Racehl Rosenthal, Dr. Charles Swanton)
Given the prevalence of LOH events detected in the treatment-naive cohorts analyzed thus far, it may be important to consider HLA LOH when designing patient-specific immunotherapy approaches, such as tumor-infiltrating lymphocyte (TIL)–based therapies and neoantigen vaccines. For example, in every patient in the TRACERx study who exhibited HLA LOH, there were predicted neoantigens binding to the lost haplotype. This observation suggests that considering HLA LOH when identifying putative neoantigens that may elicit an effective T-cell response may improve clinical response to immunotherapy. ✦
About the Authors: Dr. Rosenthal is a scientist in the Translational Cancer Therapeutics Laboratory, University College London, United Kingdom. Prof. Swanton is a clinician scientist with the Translational Cancer Therapeutics Laboratory, the Francis Crick Institute, United Kingdom.
References:
1. Hanahan D and Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674.
2. Shukla SA, Rooney MS, Rajasagi M, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152-1158.
3. Hicklin DJ, Marincola FM, Ferrone S. HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story. Mol Med Today. 1999;5:178-186.
4. Garrido F, Ruiz-Cabello F, Aptsiauri N. Rejection versus escape: the tumor MHC dilemma. Cancer Immunol Immunother. 2017;66:259-271.
5. Campoli M, Chang CC, Ferrone S. HLA class I antigen loss, tumor immune escape and immune selection. Vaccine. 2002;20 Suppl 4:A40-A45.
6. Benitez R, Godelaine D, Lopez-Nevot MA, et al. Mutations of the beta2-microglobulin gene result in a lack of HLA class I molecules on melanoma cells of two patients immunized with MAGE peptides. Tissue Antigens. 1998;52:520-529.
7. D’Urso CM, Wang ZG, Cao Y, et al. Lack of HLA class I antigen expression by cultured melanoma cells FO-1 due to a defect in B2m gene expression. J Clin Invest. 1991;87:284-292.
8. Drake CG, Jaff ee E, Pardoll DM. Mechanisms of immune evasion by tumors. Adv Immunol. 2006; 90:51-81.
9. Koopman LA, Corver WE, van der Slik AR, Giphart MJ, Fleuren GJ. Multiple genetic alterations cause frequent and heterogeneous human histocompatibility leukocyte antigen class I loss in cervical cancer. J Exp Med. 2000;191:961-976.
10. Tran E, Robbins PF, Lu YC, et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med. 2016;375:2255-2262.
11. McGranahan N, Rosenthal R, Hiley CT, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell. 2017;171:1259- 1271 e11.
12. Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109-2121.
13. Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359(6375):582-587.